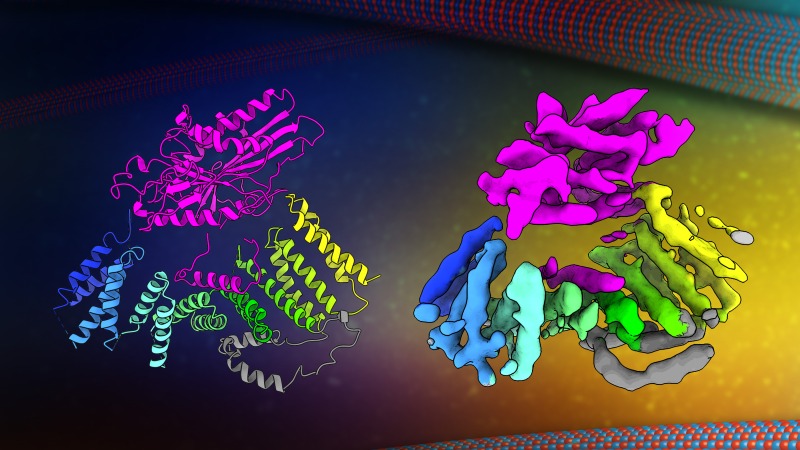
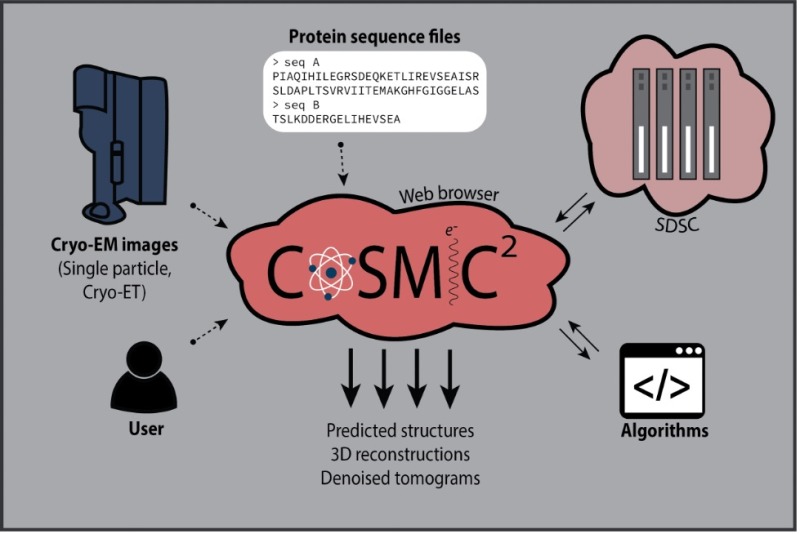
Our research team is trying to understand the molecular details determining how, where, and when motor proteins transport intracellular cargo. The past thirty years of cell biology research have set the stage for us to determine the general principles that underlie the complex process of intracellular transport.
Overarching questions that we are trying to answer:
- How is motor protein activity turned ‘on’ and ‘off’
- How do viruses hijack motor protein activity?
- How do microtubule structure and post-translational modifications affect motor protein activity?
We are approaching these questions from several angles, using cryo-electron microscopy, single molecule TIRF and biochemistry to relate protein structure to its activity in a cell.