U-M research reveals key piece of the machine that bacteria use to build useful natural products
University of Michigan researchers have discovered how the pieces of one molecular machine inside bacteria fit together, improving scientists’ abilities to recreate this well-evolved machine in the lab.
To manufacture the chemical compounds they need to thrive in their environments, bacteria and other microorganisms have evolved their own internal assembly lines — large (relatively speaking) combinations of enzymes that efficiently construct the needed compound, step by step.
With their power to speed up chemical reactions, these natural assembly lines have many potential uses in the lab, as well. One type, for example, churns out a class of compounds called polyketides, which form the basis of numerous antibiotics, immunosuppressants, cholesterol-reducers, insecticides and veterinary drugs.
But to recreate these molecular machines, and potentially improve upon them, scientists first need a detailed picture of all their pieces and how they fit together. The assembly line that builds polyketides, for instance, includes several different modules, each comprising as many as five enzymes. While previous studies have revealed what some of these pieces look like, their arrangement together remained unclear.
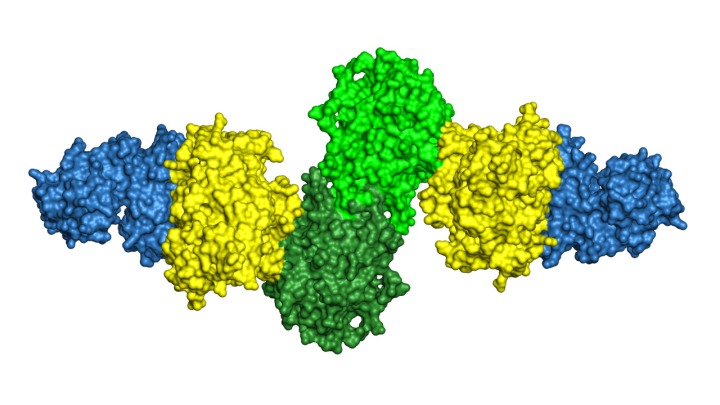
Now a new study, published June 21 in the journal Structure, offers a high-resolution picture of how the enzymes in one module fit together and interact to perform their steps in the polyketide assembly line.
“This particular module has three enzymes that catalyze reactions, one after the other,” explains Janet Smith, Ph.D., Rita Willis Professor of the Life Sciences, whose lab at the U-M Life Sciences Institute led the research. “We knew what each of them looked like. But what we didn't know — and that nobody knew — was how they are arranged when they're hooked up to each other, as they are in their natural setting. And the arrangement we saw was really surprising.”
Our findings add a new piece of the picture for these important assembly lines for natural products, and it's a piece that had never been seen before.

The team used a technique called x-ray crystallography to obtain the new structural information. This method involves converting the proteins to a crystallized form, and then beaming them with X-rays to reveal their atomic architecture. The results showed that, rather than three beads connected loosely by a string, the three enzymes are arranged tightly in a row, with the “string” holding them together like sticky tape.
“The structure of this tridomain shows how these natural product assembly lines from bacteria are distant cousins of human enzyme that makes fatty acids, and it also serves as a prototype for future engineering of these systems” says the study’s lead author Tyler McCullough, Ph.D., who recently completed his graduate work in Smith’s lab.
With a clearer picture of how the module fits together, scientists will have more opportunities not only to reconstruct it in the lab, but also to engineer new versions that can perform more reactions.
“One of the important things we can do is manipulate the DNA that codes these enzymes, to engineer pathways that can work with additional starting materials and build new products. But we first need to understand how they are put together in nature,” says Smith, who is also the Martha L. Ludwig Distinguished University Professor of Biological Chemistry in the U-M Medical School. “Our findings add a new piece of the picture for these important assembly lines for natural products, and it's a piece that had never been seen before.”
X-ray crystallography structure of three enzymes from a module of the "assembly line" that bacteria use to build polyketides, natural products with important applications in human health & welfare. The structure reveals how the three enzymes present in identical pairs fit tightly together, offering important clues for recreating this molecular machine in the lab.
Go to Article
“Structure of a modular polyketide synthase-reducing region,” Structure. DOI: 10.1016/j.str.2023.05.019


