Studying the structure of a cardiac enzyme to target heart failure
University of Michigan researchers have discovered the way in which structural features of a critical enzyme found in the cardiovascular system influence its regulation of heart activity. The findings provide new insights into a therapeutic target for treating heart disease.
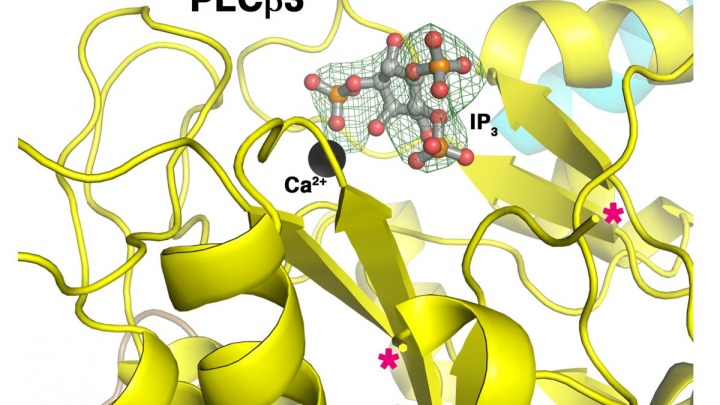
Using X-ray crystallography investigators in the John Tesmer lab at the Life Sciences Institute characterized the enzyme phospholipase C β (PLCβ), which is found in the heart and regulates a pathway that controls the heart’s pumping ability. An abnormal increase in signaling by this enzyme may lead to increased work by the heart—more contractions and greater force of contractions—which may result in permanently enlarged heart muscle cells. This enlarged heart is then unable to pump as effectively, which may lead to heart failure. The findings were published in Structure on December 2.
“When we removed specific segments of the linker, the activity of the enzyme either increased or decreased, demonstrating that each of these regions plays a unique role in regulating activity,” said Angeline Lyon, now an assistant professor of chemistry at Purdue University. “When we removed the regulatory helix, we only saw an increase in activity.”
However, the researchers did not originally predict these findings.
“It was unexpected that regions of the protein previously shown to be disordered could have such a profound negative or positive role on activity,” said Tesmer. “We were incorrect when we initially thought the regulatory helix and the linker worked together, yet this led to the more interesting finding that this structural element affects how the enzyme binds to hormone-responsive proteins that control cardiovascular function.”
“We won’t fully know how to target PLCβ therapeutically until we have a complete picture of how all of its structural components collaborate to affect its activity,” Tesmer added.
The authors next aim to study how other domains of PLCβ influence its regulatory components and how these domains bind to other proteins involved in control of its activity in cells.
John Tesmer is a research professor at the U-M Life Sciences Institute, where his laboratory is located and his research is conducted. He is also a professor of biological chemistry and pharmacology at the University of Michigan Medical School.
Go to Article
Molecular Mechanisms of Phospholipase C β3 Autoinhibition, Structure. DOI: 10.1016/j.str.2014.10.008


