Proteins are the chemical machines of living systems; they are the control, communication and molecular transportation molecules of cells, and they form part of the structural skeleton. All proteins in an organism can be identified from its genome sequence; however, the three-dimensional structure must be known to understand the function and mechanism of a protein. The overall goal of our research is to understand biological function at the molecular level through knowledge of protein three-dimensional structure.
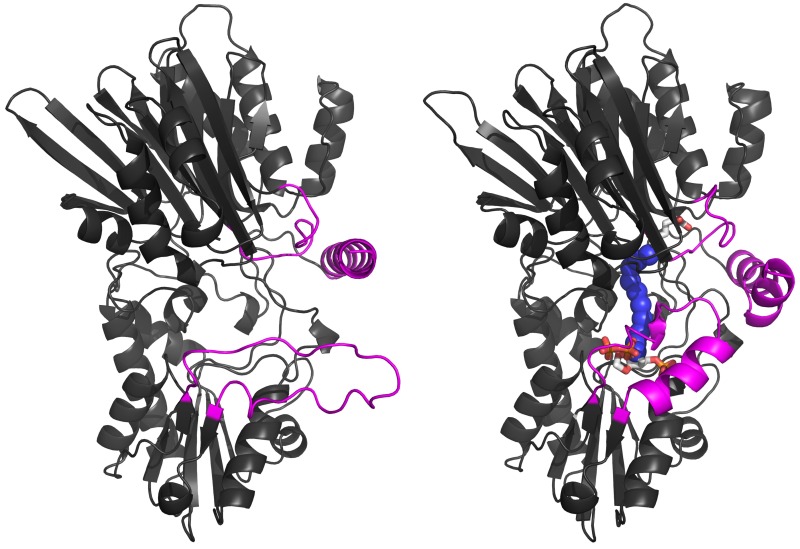
Our early studies of metabolic enzymes illustrated the sophisticated control mechanisms that balance the many biochemical pathways in living cells. The overall metabolic health of the cell influences the availability of nitrogen for synthesis of new biomolecules by using a central carbohydrate metabolite to deliver nitrogen for biosynthesis rather than a simple nitrogen molecule such as ammonia. However, cells pay a price for the homeostasis provided by such a nitrogen carrier system.
Biosynthetic pathways requiring nitrogen use "complex" enzymes to remove nitrogen from the carrier molecule glutamine. Our work elucidated the structural basis for catalysis and control in these enzymes, and has uncovered several underlying features of the relevant protein structural families.
Biosynthesis of Natural Products
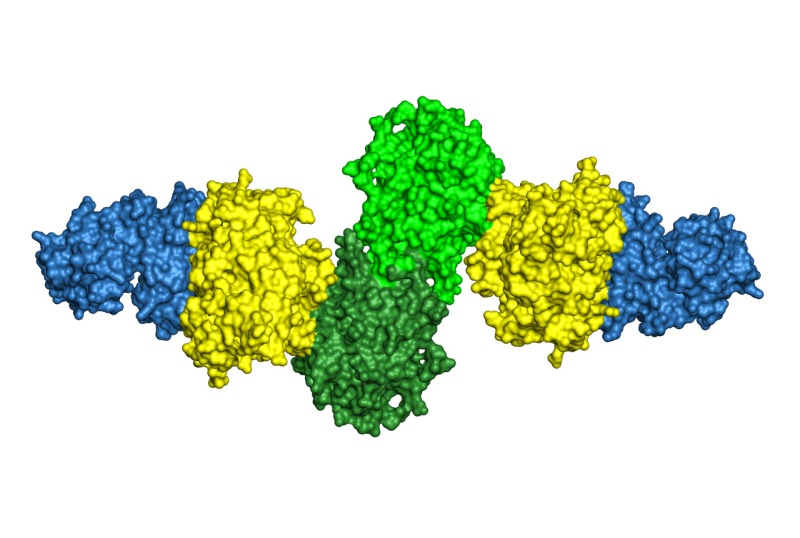
Microbes produce a plethora of secondary metabolites, “natural products”, of amazing chemical diversity. Natural products are the basis for a substantial fraction of drugs now used in the clinic, but we know from the sequences of thousands of microbial genomes that vast untapped potential exists in the earth’s biosphere. For natural product biosynthesis, Nature has adapted many enzymes of primary metabolism, such as those described above, to new catalytic functions. Our research investigates the structural basis for the fascinating catalytic potential of natural product biosynthetic enzymes.
Many natural products are synthesized by enzymes organized as biosynthetic assembly lines that are encoded in gene clusters. These systems are excellent candidates for engineering efforts to increase the diversity of bioactive compounds. We have investigated the structures of assembly line “stations” and how pathway intermediates efficiently flow from one to the next.
Antiviral Proteins
What molecular resources do we humans possess to combat viral infection? What molecular systems do viruses deploy to evade our surveillance? Our research has provided important clues about the function of several viral and host proteins involved in the host-virus cat-and-mouse game.
Flaviviruses such as dengue, Zika, West Nile and yellow fever viruses are major human pathogens. They all possess a multi-functional protein known as NS1, which assists in replication of the viral RNA genome and also aids viral spread to uninfected cells. Our structure of NS1 showed how the protein can interact with the cell surface and led to the discovery of several additional NS1 functions.


Humans have several proteins that thwart viral spread. We have studied two antiviral proteins, the restriction factor known as A3H and the zinc-finger antiviral protein ZAP. A3H binds the HIV-1 RNA genome and catalyzes hypermutation of the first DNA strand synthesized by the viral reverse transcriptase. ZAP is produced when the cell’s innate immune system detects infection by an RNA virus and traps viral RNA for destruction by a host ribonuclease. Our structures have revealed a few surprises in the RNA binding of both proteins.


