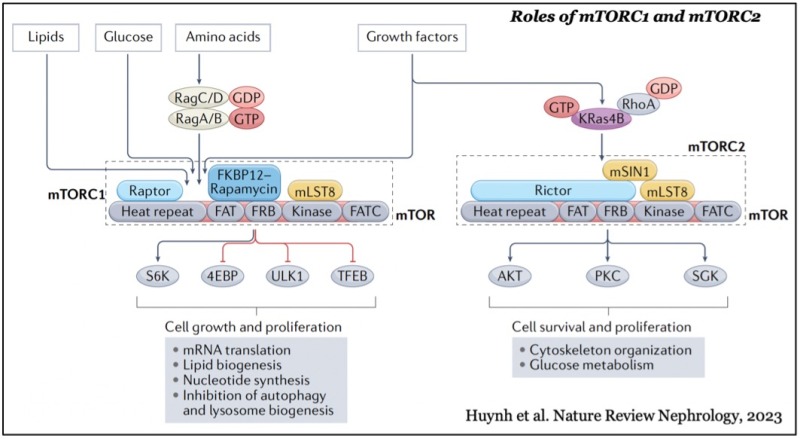
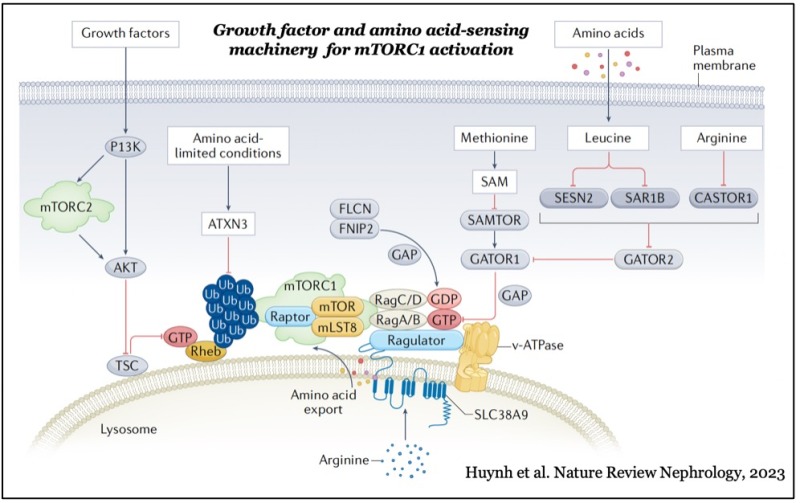
mTOR forms two distinct functional complexes — mTOR complex 1 and 2 (mTORC1 and mTORC2) — which are evolutionarily conserved. mTORC1 exists as a multiprotein complex containing mTOR, Raptor, and mLST8; mTORC2 consists of mTOR, Rictor, mSin1, and mLST8. mTORC1 can be fully activated by both growth factors and nutrients, such as amino acids and glucose on the membrane of lysosomes, a major catabolic organelle that digests unnecessary proteins and damaged organelles. Whereas mTORC1 activity is sensitive to inhibition by rapamycin, an FDA-approved anti-cancer and immunosuppressant, mTORC2 activity is resistant to the drug.
While mTORC1 stimulates major cellular anabolic processes by phosphorylating its dozens of downstream substrates, such as S6 kinase, the reason why its activation occurs on lysosomes, is not fully understood. However, recent studies suggested that mTORC1 may also sense lysosomal luminal amino acids in addition to cytosolic amino acids for its activation, highlighting that the lysosome functions as a platform for both cellular catabolism and anabolism.
In contrast, mTORC2 activity is mainly regulated by growth factors such as insulin and IGF-1 and activated at the plasma membrane. The active mTORC2 phosphorylates and activates AGC-kinases such as Akt and protein kinase C, kinases essential for cell proliferation, survival, cytoskeleton reorganization, and glucose metabolism.