Study unveils the intricate way two proteins interact to promote cell movement and metastasis
When cells in our bodies need to move — to attack an infection or heal a wound, for example — cellular proteins send and receive a cascade of signals that directs the cells to the right place at the right time. It’s a process cancer cells can hijack to spread to new tissues and organs.
Now, a team of researchers led by the University of Michigan Life Sciences Institute has shed light on a key driver of this process. The findings, published Oct. 16 in Science Advances, offer important insights into cell migration not only under normal health conditions, but also in breast, prostate, and other types of cancers.
The researchers specifically probed a protein called P-Rex1 (for phosphatidylinositol 3,4,5-triphosphate–dependent Rac exchanger 1), which is activated when it binds G protein beta-gamma. Despite the discovery of P-Rex1 more than fifteen years ago, precisely how the two proteins interact and how this interaction leads to cell movement has remained poorly understood.
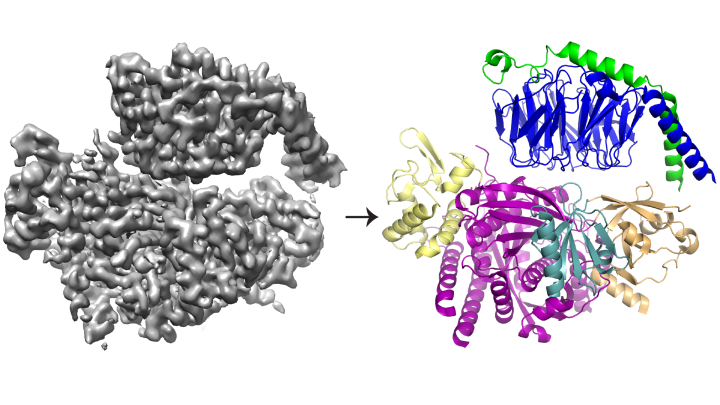
Using a combination of structural biology and biochemistry techniques, the researchers have revealed the structure of P-Rex1 bound to G beta-gamma, providing a snapshot of how this intricate activation process unfolds.

“Knowing the structure of this protein complex provides mechanistic details that allow us to understand how it is regulated,” explains Jennifer Cash, Ph.D., a researcher at the LSI and lead author of the study. “And when we understand how it’s regulated, we can start to think about how to modify that regulation and inhibit P-Rex1 signaling in cancers.”
The team found that G beta-gamma binds to an extensive surface on P-Rex1 comprised of several different protein domains. Previous studies offered conflicting arguments about which domain (or domains) bound G beta-gamma. But this latest study was able to resolve the conflict by taking a bigger-picture view of the protein, using newer technologies.
“We wanted to look at the enzyme as a whole — and to do that, we really needed to move into cryo-EM,” says John Tesmer, Ph.D., a professor of biological sciences at Purdue University and one of the study’s senior authors.
Cryo-EM, or cryo-electron microscopy, enables researchers to study proteins that cannot be visualized using other structural biology techniques — such as the complex, interwoven structure of P-Rex1. The process involves freezing proteins in a thin layer of water and then using electrons to capture images of their shape. Hundreds of thousands of images are then averaged to create a 3-D structure.
Beyond clarifying the mechanistic details of this protein-protein interaction, these new findings stand as an important touchstone in the development of cryo-EM, says Michael Cianfrocco, Ph.D., an assistant professor at the LSI and a senior author of the study.
The majority of structures determined with cryo-EM are larger, symmetric molecules. The results for P-Rex1, in contrast, represent a high-resolution structure of the smallest asymmetric protein achieved through cryo-EM to date. Furthermore, a large domain in P-Rex1 has a structure or fold that had not previously been identified in any other mammalian protein, requiring the team to build it piece-by-piece — a difficult task with cryo-EM data.
“Cryo-EM is still a relatively new field, and people are really interested in knowing how far its limits can be pushed,” says Cianfrocco, who is also an assistant professor of biological chemistry at the U-M Medical School.
Go to Article
Cryo-electron microscopy structure and analysis of the P-Rex1–G beta-gamma signaling scaffold, Science Advances. DOI: 10.1126/sciadv.aax8855


